간단한 classification
Breast Cancer Wisconsin (Diagnostic) Data Set
0. Settings
1
2
3
4
5
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
1
2
3
4
5
file_name = '/kaggle/input/breast-cancer-wisconsin-data/data.csv'
df = pd.read_csv(
file_name,
)
1. EDA
1.1. Preview
1
df.head()
| diagnosis | radius_mean | texture_mean | perimeter_mean | area_mean | smoothness_mean | compactness_mean | concavity_mean | concave points_mean | symmetry_mean | ... | radius_worst | texture_worst | perimeter_worst | area_worst | smoothness_worst | compactness_worst | concavity_worst | concave points_worst | symmetry_worst | fractal_dimension_worst | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.3001 | 0.14710 | 0.2419 | ... | 25.38 | 17.33 | 184.60 | 2019.0 | 0.1622 | 0.6656 | 0.7119 | 0.2654 | 0.4601 | 0.11890 |
| 1 | 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.0869 | 0.07017 | 0.1812 | ... | 24.99 | 23.41 | 158.80 | 1956.0 | 0.1238 | 0.1866 | 0.2416 | 0.1860 | 0.2750 | 0.08902 |
| 2 | 1 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.1974 | 0.12790 | 0.2069 | ... | 23.57 | 25.53 | 152.50 | 1709.0 | 0.1444 | 0.4245 | 0.4504 | 0.2430 | 0.3613 | 0.08758 |
| 3 | 1 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.2414 | 0.10520 | 0.2597 | ... | 14.91 | 26.50 | 98.87 | 567.7 | 0.2098 | 0.8663 | 0.6869 | 0.2575 | 0.6638 | 0.17300 |
| 4 | 1 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.1980 | 0.10430 | 0.1809 | ... | 22.54 | 16.67 | 152.20 | 1575.0 | 0.1374 | 0.2050 | 0.4000 | 0.1625 | 0.2364 | 0.07678 |
5 rows × 31 columns
1
df.info()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 569 entries, 0 to 568
Data columns (total 33 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id 569 non-null int64
1 diagnosis 569 non-null object
2 radius_mean 569 non-null float64
3 texture_mean 569 non-null float64
4 perimeter_mean 569 non-null float64
5 area_mean 569 non-null float64
6 smoothness_mean 569 non-null float64
7 compactness_mean 569 non-null float64
8 concavity_mean 569 non-null float64
9 concave points_mean 569 non-null float64
10 symmetry_mean 569 non-null float64
11 fractal_dimension_mean 569 non-null float64
12 radius_se 569 non-null float64
13 texture_se 569 non-null float64
14 perimeter_se 569 non-null float64
15 area_se 569 non-null float64
16 smoothness_se 569 non-null float64
17 compactness_se 569 non-null float64
18 concavity_se 569 non-null float64
19 concave points_se 569 non-null float64
20 symmetry_se 569 non-null float64
21 fractal_dimension_se 569 non-null float64
22 radius_worst 569 non-null float64
23 texture_worst 569 non-null float64
24 perimeter_worst 569 non-null float64
25 area_worst 569 non-null float64
26 smoothness_worst 569 non-null float64
27 compactness_worst 569 non-null float64
28 concavity_worst 569 non-null float64
29 concave points_worst 569 non-null float64
30 symmetry_worst 569 non-null float64
31 fractal_dimension_worst 569 non-null float64
32 Unnamed: 32 0 non-null float64
dtypes: float64(31), int64(1), object(1)
memory usage: 146.8+ KB
1
df.id.duplicated().sum()
1
0
1
2
# 중복된 id가 없으므로 id column을 제외하고 각 row를 독립적인 데이터로 사용
df = df.drop(['id'], axis=1)
1.1.1. 결측치 처리
1
2
# 마지막 컬럼 제외
df = df.iloc[:, :-1]
1.1.2. Label diagnosis
1
df.diagnosis.value_counts()
1
2
3
4
diagnosis
B 357
M 212
Name: count, dtype: int64
1
2
df['diagnosis'] = df['diagnosis'].apply(lambda x: 1 if x == 'M' else 0)
df.diagnosis.value_counts()
1
2
3
4
diagnosis
0 357
1 212
Name: count, dtype: int64
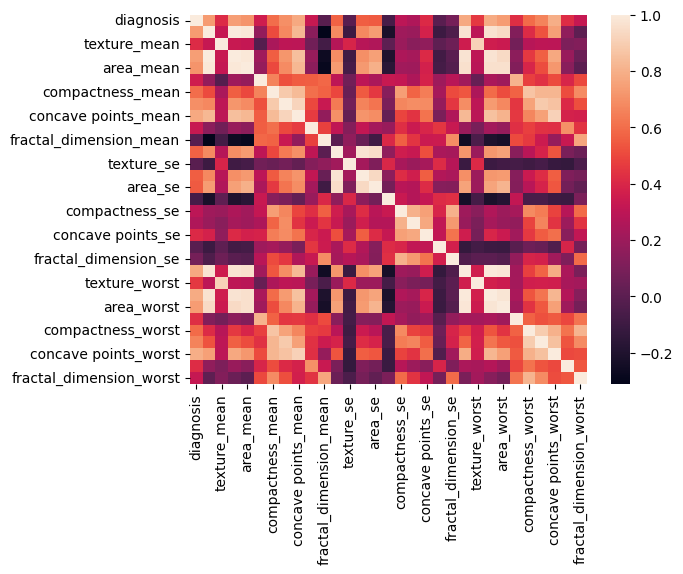
1.1.3. Correlations
1
corr = df.corr()
1
2
sns.heatmap(corr)
plt.show()
2. Predict
2.1. Data Selection
1
corr['diagnosis'].sort_values().tail(10)
1
2
3
4
5
6
7
8
9
10
11
concavity_mean 0.696360
area_mean 0.708984
radius_mean 0.730029
area_worst 0.733825
perimeter_mean 0.742636
radius_worst 0.776454
concave points_mean 0.776614
perimeter_worst 0.782914
concave points_worst 0.793566
diagnosis 1.000000
Name: diagnosis, dtype: float64
1
2
3
idx = corr['diagnosis'].sort_values().tail(10).index
X = df[idx].drop(['diagnosis'], axis=1)
y = df['diagnosis']
1
X.head()
| concavity_mean | area_mean | radius_mean | area_worst | perimeter_mean | radius_worst | concave points_mean | perimeter_worst | concave points_worst | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.3001 | 1001.0 | 17.99 | 2019.0 | 122.80 | 25.38 | 0.14710 | 184.60 | 0.2654 |
| 1 | 0.0869 | 1326.0 | 20.57 | 1956.0 | 132.90 | 24.99 | 0.07017 | 158.80 | 0.1860 |
| 2 | 0.1974 | 1203.0 | 19.69 | 1709.0 | 130.00 | 23.57 | 0.12790 | 152.50 | 0.2430 |
| 3 | 0.2414 | 386.1 | 11.42 | 567.7 | 77.58 | 14.91 | 0.10520 | 98.87 | 0.2575 |
| 4 | 0.1980 | 1297.0 | 20.29 | 1575.0 | 135.10 | 22.54 | 0.10430 | 152.20 | 0.1625 |
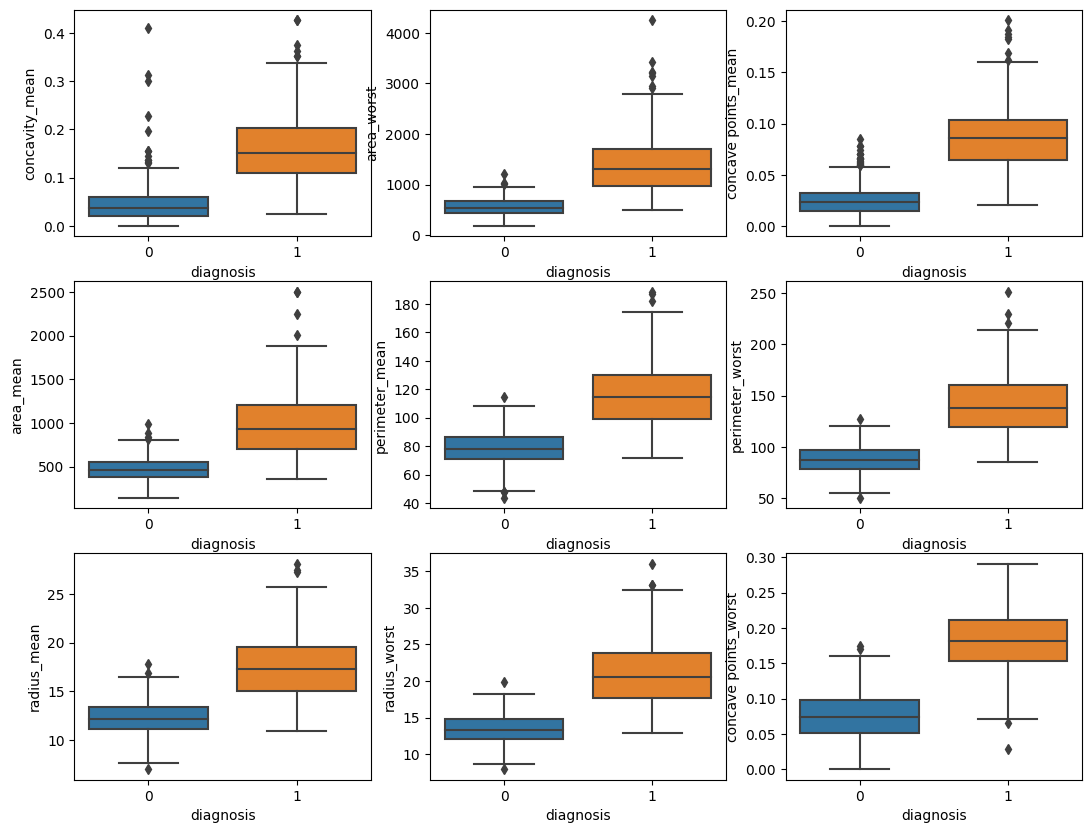
1
2
3
4
5
6
fig, axs = plt.subplots(nrows=3, ncols=3, figsize=(13, 10))
for i, col_name in enumerate(X.columns):
sns.boxplot(y=X[col_name], x=y, ax=axs[i%3][i//3])
plt.show()
2.2. Machine Learning
1
2
3
4
5
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.neighbors import KNeighborsClassifier
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split
1
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
1
2
3
4
pipe = Pipeline([
('scale', None),
('model', KNeighborsClassifier())
])
1
pipe.get_params()
1
2
3
4
5
6
7
8
9
10
11
12
13
{'memory': None,
'steps': [('scale', None), ('model', KNeighborsClassifier())],
'verbose': False,
'scale': None,
'model': KNeighborsClassifier(),
'model__algorithm': 'auto',
'model__leaf_size': 30,
'model__metric': 'minkowski',
'model__metric_params': None,
'model__n_jobs': None,
'model__n_neighbors': 5,
'model__p': 2,
'model__weights': 'uniform'}
1
2
3
4
5
6
7
8
9
10
11
12
13
param_grid = {
'model__n_neighbors':[5,10],
'scale': [StandardScaler(), MinMaxScaler()],
# 'model__class_weight': [{0:1, 1:v/2} for v in range(1,5)]
}
mod = GridSearchCV(
estimator=pipe,
param_grid=param_grid,
cv=3
)
1
mod.fit(X_train, y_train)
1
pd.DataFrame(mod.cv_results_).T
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| mean_fit_time | 0.004918 | 0.004554 | 0.004715 | 0.004649 |
| std_fit_time | 0.000317 | 0.000055 | 0.000029 | 0.00001 |
| mean_score_time | 0.012827 | 0.012498 | 0.012517 | 0.012951 |
| std_score_time | 0.000298 | 0.000277 | 0.000072 | 0.000264 |
| param_model__n_neighbors | 5 | 5 | 10 | 10 |
| param_scale | StandardScaler() | MinMaxScaler() | StandardScaler() | MinMaxScaler() |
| params | {'model__n_neighbors': 5, 'scale': StandardSca... | {'model__n_neighbors': 5, 'scale': MinMaxScale... | {'model__n_neighbors': 10, 'scale': StandardSc... | {'model__n_neighbors': 10, 'scale': MinMaxScal... |
| split0_test_score | 0.943662 | 0.929577 | 0.93662 | 0.929577 |
| split1_test_score | 0.950704 | 0.93662 | 0.943662 | 0.943662 |
| split2_test_score | 0.93662 | 0.943662 | 0.93662 | 0.93662 |
| mean_test_score | 0.943662 | 0.93662 | 0.938967 | 0.93662 |
| std_test_score | 0.00575 | 0.00575 | 0.00332 | 0.00575 |
| rank_test_score | 1 | 3 | 2 | 3 |
1
mod.score(X_test, y_test)
1
0.951048951048951
This post is licensed under CC BY 4.0 by the author.